Mechanisms of ion channel regulation investigated by extensive simulation and Markov state modeling
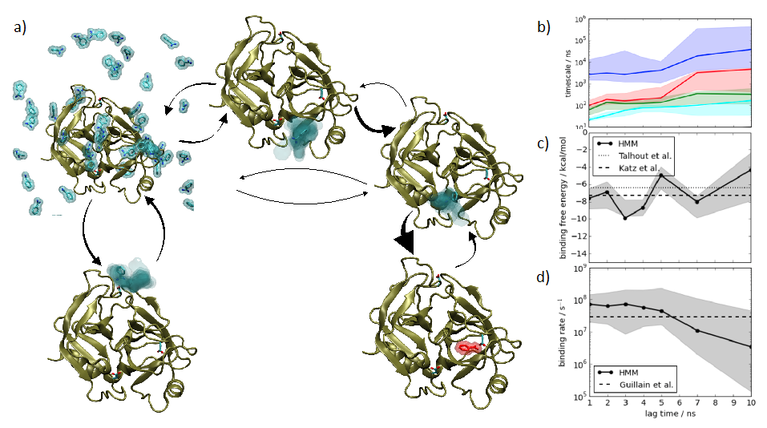
Markov state models (MSMs) are universal frameworks to encode the switching kinetics of biomolecular machines with multiple states. MSMs have been used to interpret the multi-state nature of experimental single-molecule recordings of ion channels and more recently they have been established as an inference machine to analyze high-throughput molecular dynamics (MD) simulation data. In this project, we will provide our expertise in long-timescale simulation and Markov state modeling from both experimental and simulation data to the Forschergruppe. By combining extensive GPU-driven MD simulations (100 microsecond to milliseconds) with recently developed adaptive sampling techniques and multi-ensemble estimators, we will explore the conformational dynamics of binding domains in AMPA-type glutamate receptors and their modulation by binding auxiliary proteins. Methodologically, we will develop new Bayesian inference methods to learn MSMs from single-channel data and optimally combine them with simulation data.