Combining Experiment, Simulation and Machine Learning to elucidate the activation of Glutamate receptor complexes
Glutamate receptors are tetrameric ion channels with an extended domain architecture. Recent advances in structural biology allow us to picture the interactions of AMPA-type glutamate receptors in complex with auxiliary subunits, but the dynamics of these complexes remain opaque. With this project, we aim to isolate and study the core interactions that underlie receptor activation and auxiliary protein action. The project has a methodological and an application side. On the application side, we will combine wet experiments on auxiliary protein-receptor complexes with molecular dynamics simulations to identify the targets and dynamics of the interacting loops and the mechanism of their control of channel gating. We will start with a comparison between two essential auxiliary subunits, gamma-2 (Stargazin) and gamma-8, for which structural information recently became available. We will ask if limited interactions with only two receptor subunits enough for modulation, and determine the hierarchy of positive and negative modulatory actions.
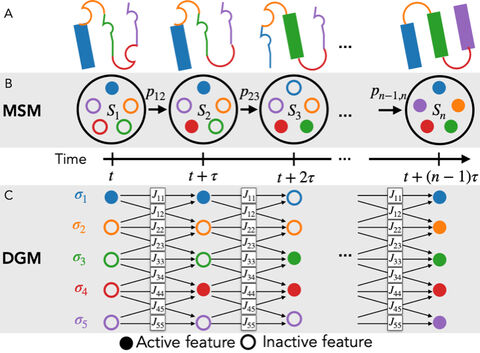
We previously used machine learning approaches to identify a conformational space in which we have built a robust MSM which allows us to estimate the kinetics of channel gating. As the multi-protein system comprised of AMPA and its auxiliary proteins is currently beyond reach of extensive MD sampling, we are compelled to further develop our machine learning methodology in the spirit of divide-and-conquering large MD systems as an assembly of metastably switching subunits. To this end we will continue and extend our VAMPnet approach. We will also pursue the application of our newly-proposed Boltzmann generators for efficiently computing free energy differences of different structures of receptor and auxiliary proteins systems. We aim to bring these methodological advances to bear on experimental observations, and pursue an integrated mechanistic model of the functional AMPA receptor states and their kinetics.
Prof. Dr. Frank Noé
14195 Berlin
Prof. Dr. Andrew Plested
Humboldt-Universität zu Berlin
Lebenswissenschaftliche Fakultät
Institut für Biologie
Invalidenstraße 110
10115 Berlin