Prof. Dr. Rainer König
Systembiologie der Sepsis/König Lab, CSCC
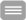

Most often, the disease or cellular condition under study has been investigated in some related
context by others having performed transcritional profiling or other sequencing based investigations
(genomics, ChIP-seq, epigenomics), Crispr/Cas9 or RNAI knockout/down screens or protein mass
spectrometry. According to the FAIR principles (Findable, Accessible, Interoperable, Reusable), such
data should be accessible, and, indeed this is often the case. Interrogating such data may be a useful
shortcut to bring up or confirm own hypotheses. However, having a bioinformatician at hand may
turn out to be the bottleneck. Hence, learning the basics for analysing such omics data can be useful
also for experimentally working biomedical scientists!
We will introduce you into the R programming language. You will learn how to write small computer
programs enabling to download such omics data and apply basic analysis and statistical methods.
Exemplarily, we will download transcriptomics data, e.g. from an infectious disease, such as from
COVID19 patients or a disease caused by another pathogen. The data will be normalized,
differentially expressed genes identified, genes set enrichment tests performed to elucidate
pathways being affected by the disease, clustering to find similarly regulated genes or samples (from
e.g. patients) and classification performed to construct a diagnosis tool.
You do not need any prior knowledge in programming and will learn how to use such an analysis
pipeline from scratch.