The era of personalized medicine promises data-driven therapeutic strategies that are tailored to individual patients based on molecular profiles. From a functional perspective, proteins are the key drivers of our cells, which also makes them primary targets of molecular therapies. Collectively, they build the proteome, which orchestrates virtually any biochemical process from energy conversion to signal transduction and the cell cycle. Conversely, aberrant protein homeostasis and signaling are hallmarks of many diseases, including cancers.
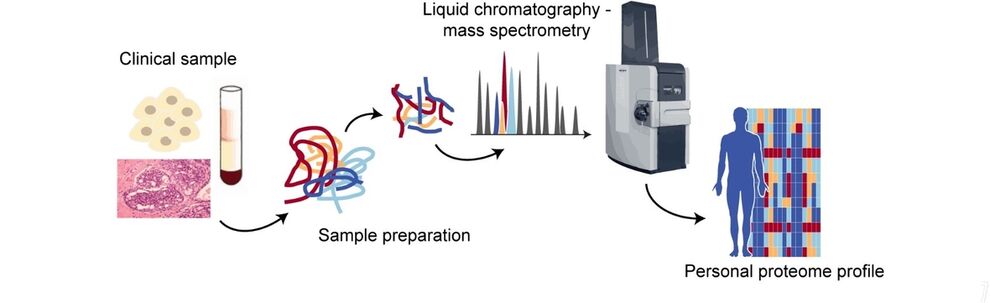
The goal of proteomics is to study the proteome in its entirety and in a quantitative, unbiased manner. Mass spectrometry (MS)-based proteomics has evolved as the method of choice for many applications. It has proven successful in the stratification of cancer subtypes from tissue biopsies as well as in disentangling cascades of post-translational modifications in signaling networks. This has been enabled by tremendous advances in the proteomics technology, which remains a driving force as the field strives towards analyzing large cohorts of clinical sample and single cells.
Our research focuses on new techniques in mass spectrometry to increase throughput, sensitivity and selectivity of the analysis. We employ latest-generation trapped ion mobility – time-of-flight mass spectrometry to analyze proteins and post-translational modifications of patient-derived cells, biofluids and tissue samples.
The summer school will introduce you to the fundamentals of MS-based proteomics. The course covers biochemical techniques to prepare samples from human specimens and process them for mass spectrometric analysis. We will make use of the latest technology to acquire quantitative data for several thousand proteins in a single experiment. Students will gain insight in different acquisition strategies and multi-dimensional separation techniques, including liquid chromatography and ion mobility spectrometry. We will use bioinformatics tools to analyze raw data and interpret the results.